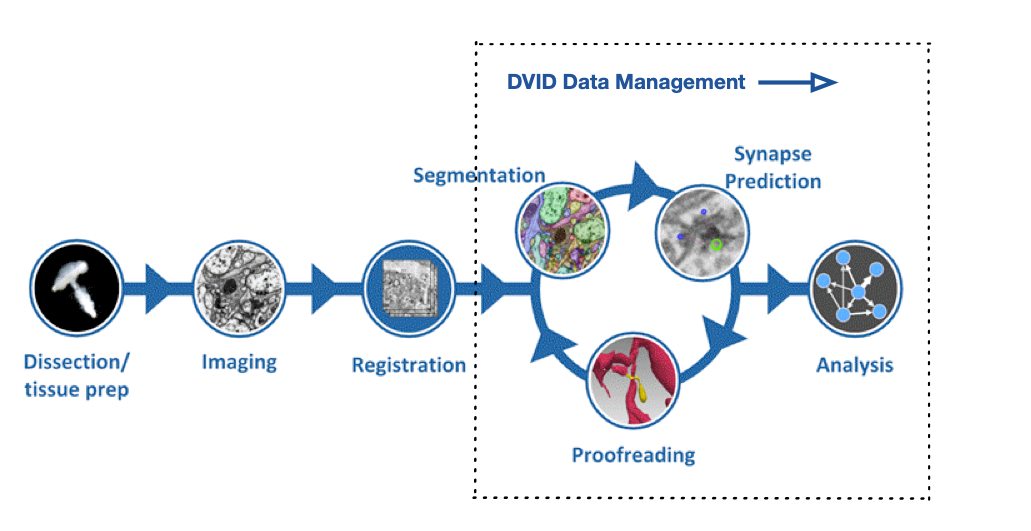
FlyEM Hemibrain V1.2 Release
Bill Katz
Senior Engineer @ Janelia Research CampusVersion 1.2 of the Hemibrain dataset has been released. We suggest you familiarize yourself with the dataset and tools to access it through the initial discussion in our previous V1.0 and V1.1 release posts. This post provides updated links and descriptions.
neuPrint+ tools from FlyEM Team
The neuPrint+ Explorer should be most visitors' first stop, and it has been updated with the V1.2 data and sports enhancements since June. We now designate the neuPrint ecosystem as neuPrint+ because we add intra-cellular interactions in addition to the inter-cellular data.
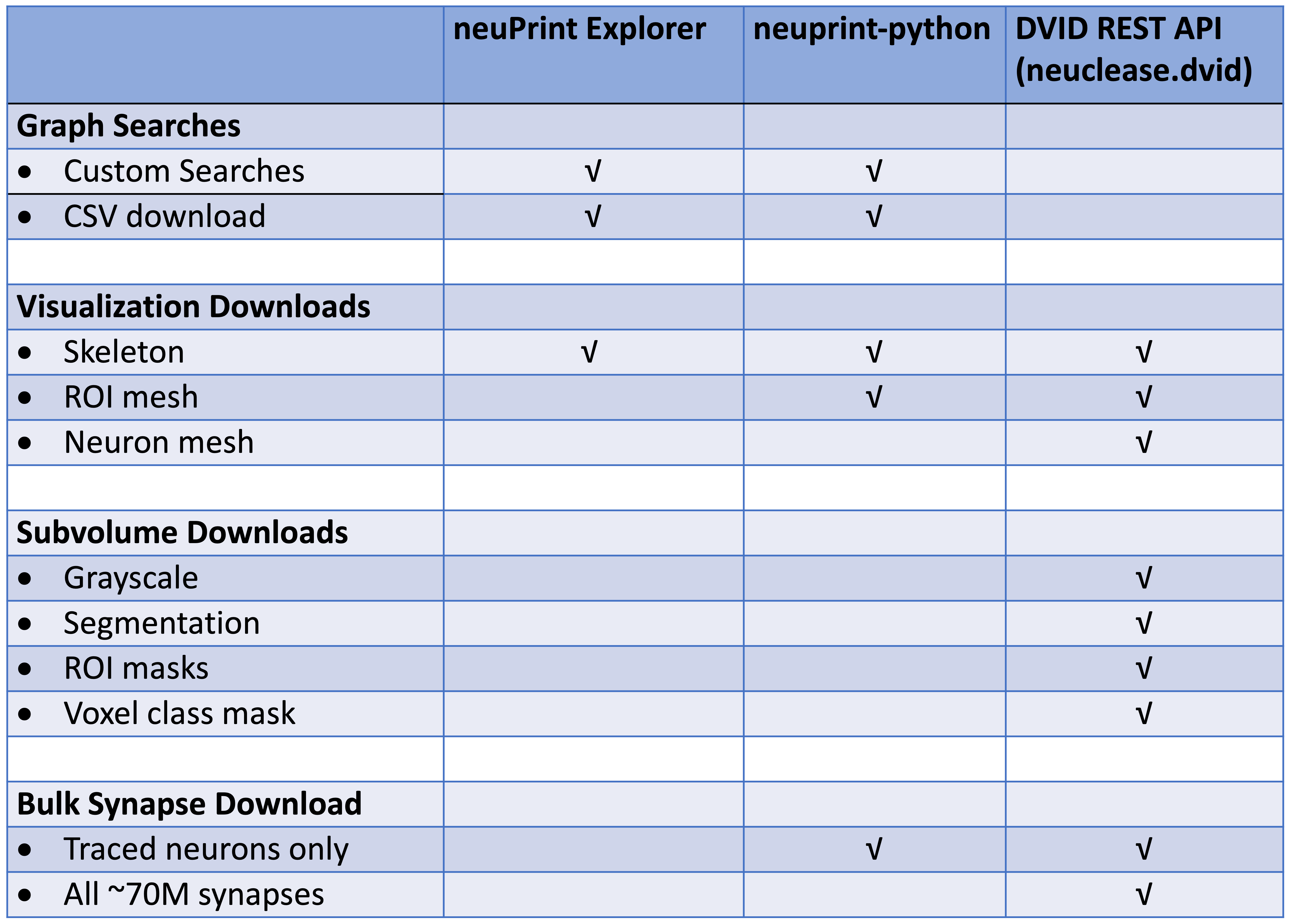
For programmatic access to the neuPrint+ database, see the neuprint-python library . For power-users who need access to the DVID database (see last section), you can try the neuclease.dvid python bindings.
This table differentiates each library based on the types of requests they support:
neuPrint tools from Collaborators
Our collaborators have developed a nice ecosystem for neurodata analysis and visualization using the neuPrint connectome service. Some of their work can be seen in their tweet thread. Code for getting and using the data include the neuprintr R library, the hemibrainr R code tailored to this Hemibrain dataset, and the NAVis python library.
Downloads
From the 26+ TB of data, we can generate a compact (44 MB) data model containing the following:
- Table of the neuron IDs, types, and instance names.
- Neuron-neuron connection table with synapse count between each pair.
- Same as above but each connection pair is split by ROI in which the synapses reside.
You can download all the data injected into neuPrint+ (excluding the 3D data and skeletons) in CSV format.
Neuroglancer Precomputed Data
Here's a link to viewing the v1.2 dataset directly in your browser with Google's Neuroglancer web app.
The hemibrain EM data and proofread reconstruction is available at the
Google Cloud Storage bucket gs://neuroglancer-janelia-flyem-hemibrain
in the Neuroglancer precomputed
format
for interactive visualization with
Neuroglancer and
programmatic access using libraries like
Cloudvolume (see below).
You can also download the data directly using the Google gsutil tool (use the -m option, e.g., gsutil -m cp -r gs://bucket mydir
for bulk transfers).
To parse the data, use one of the software libraries below or you'll have to write software to parse data using the format specification linked above.
Available data:
- EM data
- Original aligned stack (at original 8x8x8nm resolution, as well as downsamplings)
gs://neuroglancer-janelia-flyem-hemibrain/emdata/raw/jpegJPEG format - CLAHE applied over YZ cross sections (at original 8x8x8nm resolution, as well as downsamplings)
gs://neuroglancer-janelia-flyem-hemibrain/emdata/clahe_yz/jpegJPEG format
- Original aligned stack (at original 8x8x8nm resolution, as well as downsamplings)
- Segmentation
- Volumetric segmentation labels (at original 8x8x8nm resolution, as well as downsamplings)
gs://neuroglancer-janelia-flyem-hemibrain/v1.2/segmentationNeuroglancer compressed segmentation format
- Volumetric segmentation labels (at original 8x8x8nm resolution, as well as downsamplings)
- ROIs
- Volumetric ROI labels (at 16x16x16nm resolution, as well as downsamplings)
gs://neuroglancer-janelia-flyem-hemibrain/v1.2/roisNeuroglancer compressed segmentation format
- Volumetric ROI labels (at 16x16x16nm resolution, as well as downsamplings)
- Synapse detections
- Indexed spatially and by pre-synaptic and post-synaptic cell id
gs://neuroglancer-janelia-flyem-hemibrain/v1.2/synapsesNeuroglancer annotation format
- Indexed spatially and by pre-synaptic and post-synaptic cell id
- Tissue type classifications
- Volumetric labels (at original 16x16x16nm resolution, as well as downsamplings)
gs://neuroglancer-janelia-flyem-hemibrain/mask_normalized_round6Neuroglancer compressed segmentation format
- Volumetric labels (at original 16x16x16nm resolution, as well as downsamplings)
- Mitochondria detections
- Voxelwise mitochondrion class labels (at original 16x16x16nm resolution, as well as downsamplings)
gs://neuroglancer-janelia-flyem-hemibrain/v1.2/mito-classesNeuroglancer compressed segmentation format - Instance segmentation labels (at original 16x16x16nm resolution, as well as downsamplings and upsampling to 8nm)
gs://neuroglancer-janelia-flyem-hemibrain/v1.2/mito-objectsNeuroglancer compressed segmentation format - Relabeled instance segmentation, grouped by encompassing neuron ID (at original 16x16x16nm resolution, as well as downsamplings and upsampling to 8nm)
gs://neuroglancer-janelia-flyem-hemibrain/v1.2/mito-objects-grouped Neuroglancercompressed segmentation format
- Voxelwise mitochondrion class labels (at original 16x16x16nm resolution, as well as downsamplings)
Tensorstore
Earlier this year, Google released a new library for efficiently reading and writing large multi-dimensional arrays. At this time, there are C++ and python APIs. An example of reading the hemibrain segmentation is in the Tensorstore documentation.
CloudVolume
The Seung Lab's CloudVolume python client allows you to programmatically access Neuroglancer Precomputed data. CloudVolume currently handles Precomputed images (sharded and unsharded), skeletons (sharded and unsharded) and meshes (unsharded legacy format only). It doesn't handle annotations at the moment, which are usually handled by whatever proofreading system a given lab uses.
Full Datasets with DVID
For those users who want to download and forge ahead on their own copy of our reconstruction data, we can provide a replica of our production DVID system and the full Hemibrain databases. Since we are in the process of modifying the underlying system, we suggest using one of the above approaches unless you require additional data or versions only available in the production databases. Please drop us a note if you would like access to the full data. The setup for a DVID replica is described on this page.
NeuTu
NeuTu is a proofreading workhorse for the FlyEM team and can be used to proofread with DVID. It allows users to observe segmentation and to split or merge bodies if necessary. It also permits annotation, ROI creation, and many other features. Please visit this NeuTu documentation page for how to setup DVID with the Hemibrain dataset as described below.